Readme
Note
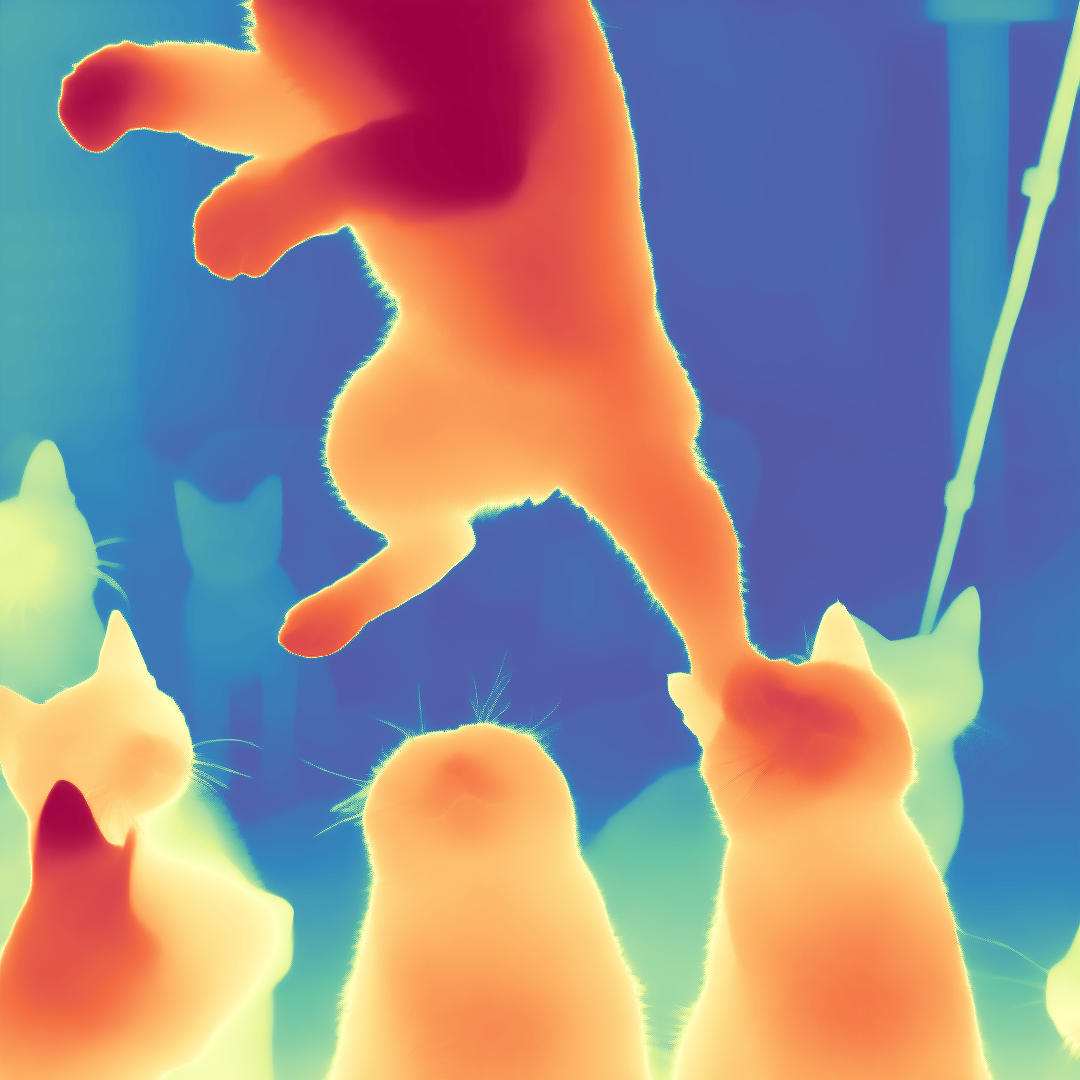
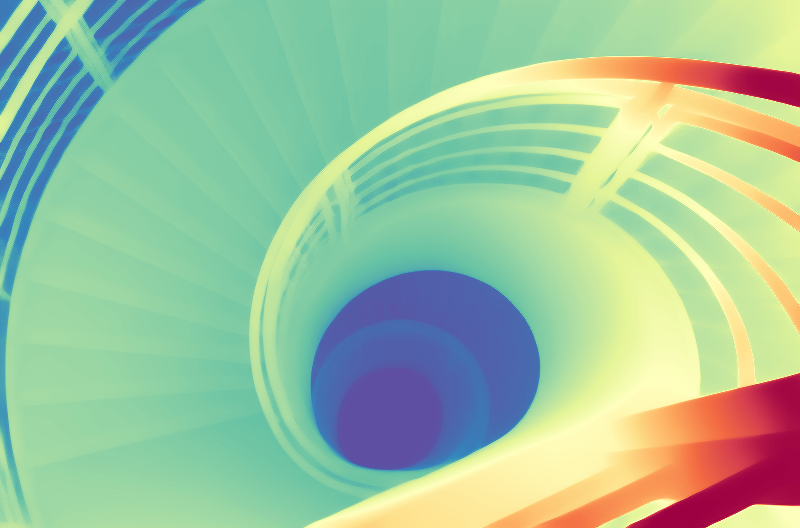
This is the jingheya/lotus-depth-d-v2-0-disparity model, which is designed for depth (disparity) regression tasks. Compared to the previous version, this model is trained in disparity space (inverse depth), achieving better performance and more stable video depth estimation.
 Lotus: Diffusion-based Visual Foundation Model for High-quality Dense Prediction
Lotus: Diffusion-based Visual Foundation Model for High-quality Dense Prediction
Authors
Jing He¹,
Haodong Li¹,
Wei Yin²,
Yixun Liang¹,
Kaiqiang Zhou³,
Hongbo Zhang³,
Bingbing Liu³,
Ying-Cong Chen¹,⁴†
Affiliations
¹ HKUST(GZ)
² University of Adelaide
³ Noah’s Ark Lab
⁴ HKUST
* Both authors contributed equally.
† Corresponding author.
We present Lotus, a diffusion-based visual foundation model for dense geometry prediction. With minimal training data, Lotus achieves SoTA performance in two key geometry perception tasks, i.e., zero-shot depth and normal estimation. “Avg. Rank” indicates the average ranking across all metrics, where lower values are better. Bar length represents the amount of training data used.
📢 News
- 2024-11-13: The demo now supports video depth estimation!
- 2024-11-13: The Lotus disparity models (Generative & Discriminative) are now available, which achieve better performance!
- 2024-10-06: The demos are now available (Depth & Normal). Please have a try!
- 2024-10-05: The inference code is now available!
- 2024-09-26: Paper released. Click here if you are curious about the 3D point clouds of the teaser’s depth maps!
🛠️ Setup
This installation was tested on: Ubuntu 20.04 LTS, Python 3.10, CUDA 12.3, NVIDIA A800-SXM4-80GB.
- Clone the repository (requires git):
git clone https://github.com/EnVision-Research/Lotus.git
cd Lotus
- Install dependencies (requires conda):
conda create -n lotus python=3.10 -y
conda activate lotus
pip install -r requirements.txt
🤗 Gradio Demo
- Online demo: Depth & Normal
- Local demo
- For depth estimation, run:
python app.py depth - For normal estimation, run:
python app.py normal
🕹️ Usage
Testing on your images
- Place your images in a directory, for example, under
assets/in-the-wild_example(where we have prepared several examples). - Run the inference command:
bash infer.sh.
Evaluation on benchmark datasets
- Prepare benchmark datasets:
-
For depth estimation, you can download the evaluation datasets (depth) by the following commands (referred to Marigold): ``` cd datasets/eval/depth/
wget -r -np -nH –cut-dirs=4 -R “index.html*” -P . https://share.phys.ethz.ch/~pf/bingkedata/marigold/evaluation_dataset/
`` - For **normal** estimation, you can download the [evaluation datasets (normal)](https://drive.google.com/drive/folders/1t3LMJIIrSnCGwOEf53Cyg0lkSXd3M4Hm?usp=drive_link) (dsine_eval.zip) into the pathdatasets/eval/normal/` and unzip it (referred to DSINE). -
Run the evaluation command:
bash eval.sh
Choose your model
Below are the released models and their corresponding configurations:
|CHECKPOINT_DIR |TASK_NAME |MODE |
|:–:|:–:|:–:|
| jingheya/lotus-depth-g-v1-0 | depth| generation|
| jingheya/lotus-depth-d-v1-0 | depth|regression |
| jingheya/lotus-depth-g-v2-0-disparity | depth (disparity)| generation|
| jingheya/lotus-depth-d-v2-0-disparity | depth (disparity)|regression |
| jingheya/lotus-normal-g-v1-0 |normal | generation |
| jingheya/lotus-normal-d-v1-0 |normal |regression |
🎓 Citation
If you find our work useful in your research, please consider citing our paper:
@article{he2024lotus,
title={Lotus: Diffusion-based Visual Foundation Model for High-quality Dense Prediction},
author={He, Jing and Li, Haodong and Yin, Wei and Liang, Yixun and Li, Leheng and Zhou, Kaiqiang and Liu, Hongbo and Liu, Bingbing and Chen, Ying-Cong},
journal={arXiv preprint arXiv:2409.18124},
year={2024}
}



-yellow)
-yellow)
